Open3DQSAR for Linux 2.24 freeware
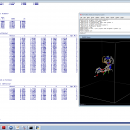
Open3DQSAR is an open-source tool aimed at pharmacophore exploration by high-throughput chemometric analysis of molecular interaction fields (MIFs). Open3DQSAR can generate steric potential, electron density and MM/QM electrostatic potential fields; furthermore, it can import GRIDKONT binary files produced by GRID and CoMFA/CoMSIA fields (exported from SYBYL with the aid of a small SPL script).
| Author | Paolo Tosco |
| Released | 2013-06-20 |
| Filesize | 1.40 MB |
| Downloads | 562 |
| OS | Linux |
| Installation | Instal And Uninstall |
| Keywords | chemometric analysis, analyze MIF, pharmacophore exploration, MIF, analysis, chemometry |
| Users' rating (15 rating) |
Open3DQSAR for Linux Free Download - we do not host any Open3DQSAR for Linux torrent files or links of Open3DQSAR for Linux on rapidshare.com, depositfiles.com, megaupload.com etc. All Open3DQSAR for Linux download links are direct Open3DQSAR for Linux download from publisher site or their selected mirrors.
| 2.24 | Jun 20, 2013 | New Release | · A bug which caused abrupt program exit when PyMOL was closed while Open3DQSAR was still running was fixed · Linking against libminizip is now optional |
| 2.23 | Jan 25, 2013 | New Release | · A bug in the import type=DEPENDENT keyword which caused an error message whenever attempting to import multiple dependent variables was fixed |
| 2.22 | Jan 4, 2013 | New Release | · The new type=MOE_GRID parameter was added to the prepare keyword to allow preparing a shape vector for the computation of interaction potentials within MOE using the same grid box settings as defined in Open3DQSAR · The new type=MOE_GRID parameter was added to the import keyword to allow importing in Open3DQSAR the interaction potential grids computed within MOE · The WriteMOEGRID.svl SVL script has been added to the util folder · Minor bugfixes all over |